Examples
A very simple example
Measuring Mg2 in the spectra number 3 to 7 of the image kenn92.fits (this image contains the atlas of galaxies published by [Kennicutt92]):
$ indexf if=kenn92.fits,3,7 index=Mg2
#
# Using indexf version: 4.1.1
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity (km/s) & error: 0, 0
#Input label file..............: undef,0,0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.1871 undef0 undef0 0.0 0.0 undef4 undef4
4 0.0485 undef0 undef0 0.0 0.0 undef4 undef4
5 0.1458 undef0 undef0 0.0 0.0 undef4 undef4
6 0.1979 undef0 undef0 0.0 0.0 undef4 undef4
7 0.2107 undef0 undef0 0.0 0.0 undef4 undef4
Note that, since each galaxy spectrum has a different radial velocity, the previous measurements are not correct (see examples below where radial velocity is properly considered).
The typical output provides a table with the measurements plus a header. The header basically gives information concerning the wavelength calibration of the data and the bandpasses defining the selected line-strength index. All the lines in this header are preceeded with a “#” symbol. It is very easy to get rid of all the lines starting with this symbol, by using:
$ indexf if=kenn92.fits,3,7 index=Mg2 verb=no
or
$ indexf if=kenn92.fits,3,7 index=Mg2 | grep -v "^#"
3 0.1871 undef0 undef0 0.0 0.0 undef4 undef4
4 0.0485 undef0 undef0 0.0 0.0 undef4 undef4
5 0.1458 undef0 undef0 0.0 0.0 undef4 undef4
6 0.1979 undef0 undef0 0.0 0.0 undef4 undef4
7 0.2107 undef0 undef0 0.0 0.0 undef4 undef4
Note that if the numbers indicating the range of spectra to be measured are missing, indexf assumes that all the spectra have to be considered
$ indexf if=kenn92.fits index=Mg2 verb=no
1 0.2149 undef0 undef0 0.0 0.0 undef4 undef4
2 0.2165 undef0 undef0 0.0 0.0 undef4 undef4
3 0.1871 undef0 undef0 0.0 0.0 undef4 undef4
4 0.0485 undef0 undef0 0.0 0.0 undef4 undef4
...
53 0.0573 undef0 undef0 0.0 0.0 undef4 undef4
54 -0.0064 undef0 undef0 0.0 0.0 undef4 undef4
55 0.0819 undef0 undef0 0.0 0.0 undef4 undef4
56 0.0310 undef0 undef0 0.0 0.0 undef4 undef4
The table with measurements contains 8 columns:
column1: spectrum number, in the range [1,NAXIS2]
column2: line-strength index (in magnitudes or Angstroms, depending on the selected index and the value of the logindex keyword in the comand line)
column3: random error in line-strength index
column4: mean signal-to-noise ratio per Angstrom in the bandpasses covered by the line-strength index
column5: radial velocity employed to measure the line-strength index
column6: radial velocity error employed to estimate the index error due to an error on radial velocity
column7: mean line-strength index in the numerical simulations performed to estimate the error due to the radial velocity error
column8: error in the line-strength index due to the radial velocity error
Using an associated image with random errors
In the case of the existence of an error FITS frame corresponding to the unbiased standar deviation of the input FITS spectra, it is straighforward to obtain the random errors associated to each line-strength index measurement:
$ indexf if=kenn92.fits,3,7 index=Mg2 ief=kenn92e.fits
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#Error FITS file...............: kenn92e.fits
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity (km/s) & error: 0, 0
#Input label file..............: undef,0,0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.1871 0.0136 14.14 0.0 0.0 undef4 undef4
4 0.0485 0.0135 14.14 0.0 0.0 undef4 undef4
5 0.1458 0.0136 14.14 0.0 0.0 undef4 undef4
6 0.1979 0.0136 14.14 0.0 0.0 undef4 undef4
7 0.2107 0.0136 14.14 0.0 0.0 undef4 undef4
In this example we have employed a scaled version of the original FITS image to simulate an associated error frame (kenn92e.fits). For this reason the signal-to-noise per Angstrom is constant. The random errors are the same in all the spectra because in the case of molecular indices (like Mg2) these errors are quite independent of absolute value of the indices. As shown in [Cardiel+98], the error on a molecular index can be estimated as
σ[Index]≈c3/SN(A),
where c3 is a parameter that depends on the wavelength limits defining the index bandpasses —which is also computed by indexf and shown in the header of the program output—, and SN(A) is the signal-to-noise per Angstrom; similarly, the c1 and c2 parameters, also given in the program output, can be employed to estimate random errors in atomic indices using
σ[Index]≈(c1−c2×Index)/SN(A)
Estimating signal-to-noise ratios
It is possible to estimate the signal-to-noise ratio using for that purpose the measurement of the standard deviation of the flux in a region relatively free from spectral features. Before measuring the standard devitation, it can be useful to subtract the local continuum by fitting a low-order polynomial. For example, we can be interested in estimating the S/N ratio in the region ranging from 5400.0 to 5500.0 Angstrom, measuring the standard deviation after subtracting the fit to a polynomial of degree 1. We can write an auxiliary file called sample.snf containing the single line:
5400.0 5500.0 1
where the first two (float) numbers provide the wavelength limits, and the third (integer) number indicates the polynomial degree. Once this file is available, one can execute:
$ indexf if=kenn92.fits,3,5 index=Mg2 snf=sample.snf
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 5
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity (km/s) & error: 0, 0
#Input label file..............: undef,0,0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.1871 0.0072 26.83 0.0 0.0 undef4 undef4
4 0.0485 0.0079 24.19 0.0 0.0 undef4 undef4
5 0.1458 0.0095 20.27 0.0 0.0 undef4 undef4
The file sample.snf can contain as many regions as desired (each region must be specified in a different line of that file). In this case, the final estimate of the S/N will be determined as the mean of the individual S/N ratios in the different regions, weighted with the number of pixels in each of those regions.
Simulating signal-to-noise ratios
The program can also estimate the random errors in each spectrum for different values of the signal-to-noise ratio per Angstrom, SN(A). In the following example we are measuring only two espectra (numbers 3 and 4), and in each of them indexf performs 5 simulations by using random SN(A) values ranging from 1 to 100 (the default values of the minmaxsn keyword):
$ indexf if=kenn92.fits,3,4 index=Mg2 nsimulsn=5
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 4
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity (km/s) & error: 0, 0
#Input label file..............: undef,0,0
#No. of simulations (S/N rat.): 5
#Minimum and Maximum S/N ratios: 1, 100
#Seed for random number........: 0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.1871 undef0 undef0 0.0 0.0 undef4 undef4
S 1 0.1993 0.0777 2.72 0.0 0.0 undef4 undef4
S 2 0.1856 0.0116 15.96 0.0 0.0 undef4 undef4
S 3 0.1907 0.0276 6.78 0.0 0.0 undef4 undef4
S 4 0.1828 0.0702 2.75 0.0 0.0 undef4 undef4
S 5 0.1876 0.0036 58.05 0.0 0.0 undef4 undef4
4 0.0485 undef0 undef0 0.0 0.0 undef4 undef4
S 1 0.0486 0.0047 40.42 0.0 0.0 undef4 undef4
S 2 0.0383 0.0850 2.10 0.0 0.0 undef4 undef4
S 3 0.0467 0.0102 16.54 0.0 0.0 undef4 undef4
S 4 0.0749 0.1562 1.25 0.0 0.0 undef4 undef4
S 5 0.0490 0.0089 19.35 0.0 0.0 undef4 undef4
Note that the measurements performed in the simulations appear in the program output preceded by the “S” character. In this particular example, after measuring the spectrum number 3, five simulations with this spectrum are carried out. Then, the spectrum number 4 is measured, and finally five simulations with this last spectrum are performed.
Since we have not fixed any value of nseed for the seed of the random number generator, the results displayed for the simulated spectra will change every time the last command is executed.
Using a fixed radial velocity
In the first example we were using a (by default) radial velocity of 0 km/s. In the following example we are using a fixed radial velocity of 1500 km/s (the same for all the measured spectra) and, simultaneously, we ask the program to estimate the effect of a random error in this velocity of 200 km/s:
$ indexf if=kenn92.fits,3,7 index=Mg2 rv=1500,200
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity (km/s) & error: 1500, 200
#No. simulations for err_rvel..: 100
#Input label file..............: undef,0,0
#Seed for random number........: 0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.2384 undef0 undef0 1500.0 200.0 0.2382 0.0014
4 0.0713 undef0 undef0 1500.0 200.0 0.0702 0.0015
5 0.1836 undef0 undef0 1500.0 200.0 0.1850 0.0036
6 0.2142 undef0 undef0 1500.0 200.0 0.2124 0.0090
7 0.2733 undef0 undef0 1500.0 200.0 0.2719 0.0047
Since we have not fixed any value of nseed for the seed of the random number generator, the results displayed in columns number 7 and 8 will change every time the last command is executed.
Using variable radial velocities
Since in a normal situation it is very likely that each spectrum may exhibit a different radial velocity (e.g. due to the presence of rotation curve in a given object), we can indicate the actual radial velocity for each individual espectrum from an external ASCII file. In the following example, the velocities are stored in a file called radvel.dat). Within this file, the radial velocities are found in the first data column, whereas the second column indicates that these velocities have an associated error of 350 km/s:
$ indexf if=kenn92.fits,3,7 index=Mg2 rvf=radvel.dat,1,2
#
# Using indexf version: x.x.x
#
WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity & error from..: radvel.dat,1,2
#No. of simulations (Rvel.err): 100
#Input label file..............: undef,0,0
#Seed for random number........: 0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.2377 undef0 undef0 1640.0 350.0 0.2343 0.0099
4 0.2899 undef0 undef0 6480.0 350.0 0.2852 0.0089
5 0.1896 undef0 undef0 1210.0 350.0 0.1858 0.0060
6 0.2263 undef0 undef0 940.0 350.0 0.2217 0.0054
7 0.2756 undef0 undef0 1250.0 350.0 0.2702 0.0090
Appending additional information in the program output
A very useful feature of indexf is the possibility of appending additional information next to the measurements of each spectrum in the program output. For example, if one wants to append the information contained in the file radvel.dat (in the range comprised between the character number 18 and 32), the instruction to be executed is:
$ indexf if=kenn92.fits,3,7 index=Mg2 rvf=radvel.dat,1,2 ilabfile=radvel.dat,18,32
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Mg2
# <code>: 1 (molecular index)
# <blue bandpass>: 4895.12 4957.62
# <central bandpass>: 5154.12 5196.62
# <red bandpass>: 5301.12 5366.12
#c1 (error estimation from S/N): 7.56651
#c2 (error estimation from S/N): 0.178035
#c3 (error estimation from S/N): 0.193299
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity & error from..: radvel.dat,1,2
#No. of simulations (Rvel.err): 100
#Input label file..............: radvel.dat,18,32
#Seed for random number........: 0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (mag) (mag) per ang (km/s) (km/s) (mag) (mag)
#====== ======= ======== ======= ======== ======== ======== ========
3 0.2377 undef0 undef0 1640.0 350.0 0.2331 0.0109 NGC4648 E3
4 0.2899 undef0 undef0 6480.0 350.0 0.2850 0.0071 NGC4889 E4
5 0.1896 undef0 undef0 1210.0 350.0 0.1859 0.0066 NGC3245 S0
6 0.2263 undef0 undef0 940.0 350.0 0.2217 0.0051 NGC3941 SB0/a
7 0.2756 undef0 undef0 1250.0 350.0 0.2703 0.0078 NGC4262 SB0
Since all the lines starting by the “#” character in the ilabfile are ignored, it is also possible to concatenate the results of different executions of indexf (note that the header of the program output is constituted by lines starting by “#”). For example:
$ indexf if=kenn92.fits,0,0 index=Mg2 rvf=radvel.dat,1,2 > kenn92_Mg2.out
$ indexf if=kenn92.fits,3,7 index=Hbeta rvf=radvel.dat,1,2 ilabfile=kenn92_Mg2.out,11,16
The first execution of indexf is employed to measure Mg2 in all the spectra of the file kenn92.fits, and the information is sent to the output file kenn92_Mg2.out. The second execution of the program makes use of the file just created to append the Mg2 measurements to the 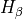 determinations.
determinations.
#
# Using indexf version: x.x.x
#
#WARNING: keyword CTYPE1 not found. Assuming CTYPE1=WAVE
#WARNING: keyword CUNIT1 not found. Assuming CUNIT1=Angstrom
#WARNING: keyword CRPIX1 not found. Assuming CRPIX1=1
#------------------------------------------------------------------------------
#Index.........................: Hbeta
# <code>: 2 (atomic index)
# <blue bandpass>: 4827.88 4847.88
# <central bandpass>: 4847.88 4876.62
# <red bandpass>: 4876.62 4891.62
#c1 (error estimation from S/N): 7.30074
#c2 (error estimation from S/N): 0.253939
#c3 (error estimation from S/N): 0.275711
#Input FITS file...............: kenn92.fits
# <NAXIS1>: 1726
# <NAXIS2>: 56
# <OBJECT>: KENNICUTT 1992
#First & last spectrum.........: 3, 7
#CRVAL1, CDELT1, CRPIX1........: 3650, 2, 1
#Radial velocity & error from..: radvel.dat,1,2
#No. of simulations (Rvel.err): 100
#Input label file..............: kenn92_Mg2.out,11,16
#Seed for random number........: 0
#Percentile for continuum (%)..: -1
#Boundary fit for continuum....: 0
#Systematic error (additive %).: 0
#Linearity error (power law)...: 0
#Flux scale factor.............: 1
#------------------------------------------------------------------------------
#
#spect. index err_phot S/N RVel. RVel.err ind_rvel err_rvel
#number (ang) (ang) per ang (km/s) (km/s) (ang) (ang)
#====== ======= ======== ======= ======== ======== ======== ========
3 1.6123 undef0 undef0 1640.0 350.0 1.4959 0.2382 0.2377
4 1.1281 undef0 undef0 6480.0 350.0 1.0917 0.4011 0.2899
5 1.6174 undef0 undef0 1210.0 350.0 1.4826 0.4643 0.1896
6 1.8630 undef0 undef0 940.0 350.0 1.7532 0.4480 0.2263
7 1.1264 undef0 undef0 1250.0 350.0 1.0286 0.3768 0.2756
Note that since the ilabfile must contain NAXIS2 lines that do not start by the “#” character (and any arbitrary number of lines starting by “#” which are ignored), the execution of indexf to create the file kenn92_Mg2.out was performed asking the program to measure the full set of spectra. However, in the second execution we do not need to measure again the full frame.
Using indexf in scripts
It is useful to employ script files to execute indexf. For example, if we have a script file called run_indexf.sh containing:
#!/bin/bash
indexf \
if = kenn92.fits,0,0 \
index = Mg2 \
ief = kenn92e.fits \
rvf = radvel.dat,1,2 > kenn92_Mg2.out
#
indexf \
if = kenn92.fits,0,0 \
index = Hbeta \
ief = kenn92e.fits \
rvf = radvel.dat,1,2 \
ilabfile = kenn92_Mg2.out,11,16 > kenn92_Hbeta_Mg2.out
after giving execution permission to this file (e.g. $ chmod u+x run_indexf.sh) the instruction
$ ./run_indexf.sh
will run indexf twice and generate the output files kenn92_Mg2.out and kenn92_Hbeta_Mg2.out.
Plotting
Plots with indexf are controlled by the keywords grdev and by plotmode (remember that plots are available only if the program has been installed with PGPLOT; see installation instructions above).
By default, grdev is set to /Xserve, which allows the user to display the result instantly on the screen:
$ indexf if=kenn92.fits,3,10 index=Mg2 ief=kenn92e.fits rvf=radvel.dat,1,2 plotmode=-1
where having used plotmode =-1 determines that the user must hit return to display the different spectra.
It is also possible to save the graphic output into a file. For example, the following instruction:
$ indexf if=kenn92.fits,3,10 index=Mg2 ief=kenn92e.fits rvf=radvel.dat,1,2 \
plotmode=-2 grdev=plot.ps/cps nwindows=2,2 > kenn92_Mg2.out
generates a color postscript file called plot.ps (a PDF version of this file is available here), containing 2 pages with 4 panels/page. The spectra are plotted with error bars. The program output is sent to the file kenn92_Mg2.out.