Using the program
With the aim of facilitating the inclusion of this program in scripts, indexf has been written assuming that all the information is transferred to the program through the command line via pairs keyword=keyvalue:
$ indexf keyword1=keyval1 keyword2=keyval2 ...
A full list of the available keyword=keyvalue pairs (as well as the list of line-strength indices defined in the file indexdef.dat) can be easily obtained just by typing the name of the program without any argument:
$ indexf
#
#Using indexf version: 4.2.2 (PGPLOT enabled)
#
#Index definitions..: /usr/local/share/indexf/indexdef.dat
#Keyword definitions: /usr/local/share/indexf/inputcl.dat
#
Usage:
$ indexf keyword1=keyval1 keyword2=keyval2 ...
where:
default
keyword keyvalues keyval description
======= ========= ===========================================================
if undef,0,0 #input file name,first spectrum,last spectrum (0,0=all)
index undef #index identification name
ief undef #input error file (incompatible with "snguess")
contperc -1 #if greater than zero, indicates percentile for continuum
boundfit 0 #if greater than zero, use boundary fit for continuum
snf undef #S/N estimation file (incompatible with "ief")
rv 0,0 #radial velocity (km/s),radial velocity error
rvf undef,1,0 #radial velocity file name, column data, column error
vacuum 0 #wavelength scale (0=air, 1=vacuum1, 2=vacuum2,...)
nsimul 100 #number of simulations to estimate radial velocity error
logindex no #measure indices in logarithmic units
verb yes #display intermediate information
nsimulsn 0 #number of simulations with different S/N ratios
minmaxsn 1.0,100.0 #minimum,maximum S/N ratios per Ang. in simulations
ilabfile undef,0,0 #input label file (first and last character (0,0=all)
biaserr 0 #systematic error (additive % of the continuum level)
linearerr 0 #linearity error: flux=flux_original^(1+linearerr)
plotmode 0 #plot mode (0:none; 1:pause, 2:no pause; -:with error bars)
grdev /Xserve #PGPLOT graphic device
nwindows 1,1 #number of panels (NX,NY) in the plotting device
plottype 1 #0: simple plot; >0: plot with additional information
nseed 0 #seed for random numbers (0=use computer time)
fscale 1.0 #flux scale factor (measured spectrum = original/fscale)
pyndexf no #echo data for communication with pyndexf (python script)
> Molecular indices: CN1 CN2 HgVA125 HgVA200 HgVA275 Mg1 Mg2 TiO1 TiO2
> Atomic indices: OII3727 HdA HdF Ca4227 G4300 HgA HgF Fe4383 Ca4455 Fe4531
C4668 Hbeta Hbeta_p OIII_1 OIII_2 OIII_2ne Fe5015 Mgb5177 Fe5270 Fe5335
Fe5406 Fe5709 Fe5782 Na5895 NaI_K FeIA_K FeIB_K CaI_K MgI_K
> D4000 like indices: D4000
> B4000 like indices: B4000
> Color like indices: CO_KH
> Emission lines: OII3727e
> Generic discontinuities: D_CO Frog_ind
> Generic indices: CaT PaT CaT_star CaT1 CaT2 CaT3 CaZHOU CaDH CaJON MgI FeI
Puxley Frog_gen
> Slope indices: sTiO
The following list gives a more detailed description of the keywords:
- if=<str,int,int>
Input FITS file name,n1,n2.
NAXIS1andNAXIS2are assumed to correspond to the spectral and spatial direction, respectively. The FITS header must contain the information concerning the wavelength calibration (CRVAL1,CDELT1,CRPIX1; if not present,CTYPE1=WAVEandCUNIT1=Angstromare assumed). If the input file contains more than 1 spectrum (NAXIS2> 1), all the spectra are measured using the sameCRVAL1,CDELT1,CRPIX1,…The two integers after the file name indicate the first and last spectrum to be measured. If n1=n2=0 (or if no numbers are provided) all the spectra are measured (i.e., n1=1 and n2=``NAXIS2`` are used).
Mandatory: yes
Default: undef,0,0
- index=<str>
Name of the line-strength index to be measured. The whole set of indices is defined in the file indexdef.dat. It is possible to obtain a list with all the defined indices by executing indexf without any argument in the command line, i.e.
$ indexf
Mandatory: yes
Default: undef
- ief=<str>
Input FITS file name containing the error spectra (unbiased standard deviation). The FITS header must contain the same information concerning the wavelength calibration than the data FITS file. If this file is undef, no error computation is performed. The FITS images containing the spectra and their associated error spectra must also have the same dimensions.
Mandatory: no
Default: undef
- contperc=<int>
Option under development; still not available.
Mandatory: no
Default: -1
- boundfit=<int>
Option under development; still not available.
Mandatory: no
Default: 0
- snf=<str>
S/N estimation file (incompatible with the keyword
ief). When this option is selected, the user can use an external ascii file containing different lines, each one containing 3 numbers:wavelength1 wavelength2 poldegree
Each line defines a wavelength region, ranging from wavelength1 to wavelength2, where the program fits a polynomial of degree given by poldegree. Note that the three numbers are not separated by commas, and that wavelength1 and wavelength2 must be floats, whereas poldegree must be an integer. This fit is subtracted from the original spectrum, and the standard deviation is computed in the residuals. This r.m.s. is used as an estimate of the S/N ratio. Then the program generates a simulated error spectrum with the estimated S/N ratio prior to the index measurement. Note that when several regions are used, an averaged S/N is employed (average computed weighting with the number of pixels in each wavelength region).
Mandatory: no
Default: undef
- rv=<float,float>
Radial velocity (km/s) and radial velocity error (km/s) for all the spectra in the FITS file. If these numbers are different for each spectrum, the keyword
rvfshould be used instead ofrv.Mandatory: no
Default: 0,0
- rvf=<str,int,int>
ASCII file name,n1,n2.
File containing a table with the radial velocity (km/s) and associated radial velocity error (km/s) for all the spectra in the input FITS file. n1 and n2 indicate the columns where those numbers can be found within the table. If n2=0 no errors in radial velocity are available. This ASCII file must contain a number of rows identical to the number of spectra in the input FITS file.
If the
rvfkeyword is present, the keywordrvis ignored (even if present in the command line).The lines starting by the character “#” in this ASCII file are ignored. Anyway, the total number of lines that do not start by the “#” character must be equal to
NAXIS2(i.e., there must be a valid radial velocity for each spectrum).Mandatory: no
Default: undef,1,0
- vacuum=<int>
Integer number indicating whether the wavelength calibration of the spectra is given in air (default
vacuum= 0), or in vacuum (integer 1,2,3). Note that for most line-strength indices wavelengths in indexdef.dat are given in the air (exceptions are near-IR indices, like D_CO, which bandpasses are expressed as vacuum wavelengths). If your spectra have been reduced using a wavelength calibration in vacuum, you can handle this by setting the parameter vacuum to 1, 2 or 3 when running the program:If vacuum is different from zero, the bandpasses given in indexdef.dat are transformed from air to vacuum prior to the measurement of the line-strength indices. If the wavelengths in indexdef.dat are already in vacuum, you do not need to use any vacuum correction (vacuum = 0 is fine in this case), although your spectra should have been reduced with a wavelength calibration in vacuum.
Mandatory: no
Default: 0
- nsimul=<int>
Number of simulations to estimate the effect of a radial velocity error in the measurement of the indices. In each simulation, a random radial velocity is drawn, following a Gaussian distribution around the initial value, with a dispersion given by the radial velocity error. The dispersion of the different indices derived from all the simulations is used as the estimation of the error.
Mandatory: no
Default: 100
- logindex=<str>
Indicates whether the indices must be measured in logarithmic units (i.e., atomic indices are measured like the molecular indices), following [Colless+99].
Mandatory: no
Default: no
- verb=<str>
Display additional information in the program output (header).
Mandatory: no
Default: yes
- nsimulsn=<int>
Number of simulations to estimate random errors in the line-strength indices. This option is available only when
ief= undef. In each simulation a random signal-to-noise ratio (in the range defined by the keywordminmaxsn) is employed.When using this option, the program outputs the result of each simulation using the prefix “S” in front of each simulation number.
Mandatory: no
Default: 0
- minmaxsn=<float,float>
Minimum and maximum signal-to-noise ratios to be employed in the simulations when
nsimulsn> 0.Mandatory: no
Default: 1.0,100.0
- ilabfile=<str,int,int>
ASCII file name,n1,n2.
File containing information for each spectrum in the input FITS file. The information contained between the columns defined by the characters [#n1,#n2] is appended at the end of the output lines with the index measurements. In this way it is easy to identify which object correspond to each spectrum. If n1=n2=0 all the information in every line of the ASCII file is employed.
The lines starting by the character “#” in this ASCII file are ignored. This helps to concatenate the results of different executions of indexf.
This option only works when the ASCII file contains information for all the spectra (
NAXIS2lines that do not start by the “#” character, and any number of lines starting by “#” which are ignored). This limitation has been established in order to prevent possible mistakes when appending files (the measure of subsections of frames is prone to errors without introducing additional checks; the approach here adopted seems safer).Mandatory: no
Default: undef,0, 0
- biaserr=<float>
[This option is not yet available for all type of line-strength indices. At present only for molecular, atomic, D4000-like and B4000-like.]
Constant number (parametrized as the percentage of the flux in the continuum) that is added to the whole spectrum before measuring the index. This can option can be employed to estimate the systematic error produced by a wrong background determination (due, for example, to scattered light). Note that this option cannot be used simultaneously with
linearerr.Mandatory: no
Default: 0
- linearerr=<float>
Parameter that allows to introduce the effect of a variation in linearity. Before measuring the indices, the flux is modified following the expression:
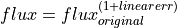 . Note that this option cannot be used simultaneously with
. Note that this option cannot be used simultaneously with biaserr.Mandatory: no
Default: 0
- plotmode=<int>
If 0 no plots are made; otherwise, plot spectra.
1: then pause between plots (you must hit return after each plot) 2: no pause between plots +n: plot spectra without error bars (even when available) -n: plot error bars (only when available)
The plots show a subsection of the spectral range containing the relevant bandpasses. The X-axis is represented with three scales: pixels (bottom axis), observed wavelength (upper top axis), and rest-frame wavelength (lower top axis). The name of the file and the spectrum number being measured is indicated in the upper right corner, whereas the index is annotated in the upper left corner.
Mandatory: no
Default: 0
- grdev=<str>
PGPLOTgraphics device. Usegrdev=’?’ to get access to the list of available devices.Mandatory: no
Default: /Xserve
- nwindows=<int,int>
Number of panels (NX,NY) in the plotting device.
Mandatory: no
Default: 1,1
- plottype=<int>
0: simple plot (single X axis and simple labels) 1: plot with additional information 2: draw different continua when simulating radial velocity errors
Mandatory: no
Default: 1
- nseed=<int>
Positive integer number used as seed for random numbers (if 0, use computer time). Using a constant seed allows to repeat simulations.
Mandatory: no
Default: 0
- fscale=<float>
Flux scale factor (measured spectrum = original/
fscale).Mandatory: no
Default: 1.0
- pyndexf=<str>
Display additional information to facilitate the execution of the python code pyndexf. It should only be activated from pyndexf.
Mandatory: no
Default: no
Note
The the pairs keyword=keyvalue can be given in any order in the command line.
The only mandatory keywords are
ifandindex(see examples below).When the keyvalue of a given keyword consists in several items separated by commas, no blank spaces can be left within those items. However, blank spaces may be left before or after the “=” sign separating the keyword from the keyvalue.
Warning and error codes
If something fails while measuring the line-strength indices (or simply some measurements cannot be completed due to the lack of information), the anomalous values are displayed with the following codes:
undef0: no error file available
undef1: something fails while estimating radial velocity errors (likely the Monte Carlo simulation has requested indexf to measure the line-strength index outside the wavelength coverage of the spectrum)
undef2: some index bandpasses are outside of the wavelength range covered by the spectrum
undef3: negative or null error values
undef4: no simulations with radial velocity error